Hello Hive Mind,
Logs with diagnosis and system health monitoring data can be used to tune the system/process perform better. One of our clients, one of the largest manufacturers of computer hardware, approached us with an interesting problem with serious business concern. Dealing with increasing, precious log data was a problem that was knocking at their doorstep. The challenging bit for us was to design a system, which can handle the scale and also allow for building models, which can analyze the collected data.
The following is a journey of how we solved a distributed Log-Processing problem by integrating various systems built around the Hadoop Eco-System. The problem at hand was to build an efficient system, which could make querying of semi-structured data more easy. This meant that the system had to handle the ever-growing data in an efficient manner by ingesting, processing and storing data into a reliable and query-able eco-system.
Hadoop is a framework that allows for the distributed processing of large data sets across clusters of computers using simple programming models. It is designed to scale up from single servers to thousands of machines, each offering local computation and storage. Rather than relying on hardware to deliver high-availability, the framework itself is designed to detect and handle failures at the application layer and hence delivering a highly available service on top of a cluster of computers, each of which may be prone to failures.
Why Hadoop?
This was one of the primary questions, which our client had, Why Hadoop? There were certain amount of constraints and the nature of data itself, which pushed us to choose Hadoop and its eco-system.
Volume of data – It was primarily tens of thousands of devices, manufactured and deployed by our client, sending diagnostic reports on a periodic basis. It could be any where in the range of several tens or hundreds of GBs of data per day. Apart from the scale issue, the core problem was that the diagnostic logs had a variety of formats. Currently they had an ETL process that stages the data and uploads to their final store for analysis. With Hadoop, since the file system does not impose any restrictions on the format of data stored, it will be easy to on-board the data and perform ETL on top of it from within Hadoop. This would allow the ETL process to be parallelized through MapReduce and made more efficient. Also, the data was sensitive and not to be made available outside of client infrastructure (Ruled out a lot of proprietary and cloud based solutions, the likes of Splunk, Big Query etc.) The other requirement was the ability to build statistical or otherwise models, which derived useful insights from the data. It was also required that the current online applications consuming the data (either for reporting purpose, monitoring purpose or for other processing) should still be able to function with minor changes and without much effort.
Summarizing all of it, it was required to build a system, which could collect data from various sources in real-time, pre-process to get it into a query-able format and in the end extract information to more traditional storage mechanisms like RDBMS for applications reporting the derived insights.
This is where Hadoop felt right at home. Reliable storage of large scale data, query using HIVE, pre-processing either using Pig or custom map-reduce jobs, complex workflow management using Oozie or Cascading or Azkaban, data ingestion using Flume or Storm or Scribe and data migration using Sqoop.
It is important to understand the practices, workflows and usage patterns commonly seen in many enterprise systems while building a custom solution. The following is a summary of our understanding with BigData applications.
The data lifecycle describes how data moves through the application from ingestion to archival. One framework we have found useful for analyzing the way the application needs to be built is to look at each phase of the lifecycle, consider what are our requirements, trade-offs, technological considerations and choices.
- Acquisition / Ingestion
This stage in the application is responsible for handling the gushing stream of input data. The solution at this stage is responsible for controlling certain scale related parameters like:
- How fast or slow do we require data to move in?
- Is it to be streamed or can we move it as bulk uploads?
- What is the unit of processing? – Generally in a temporal scope – for e.g. daily, hourly etc.
- Preparation
Conventional ETL (Extract, Transform, Load) phase. This is also the phase where we might want to consider how we will manage our schema. How we will manage changes to schema etc. Also decide what our requirements are for anonymizing data, how to handle missing values etc. The data that comes in from the acquisition stage is ‘raw’ data. After the preparatory stages, the data is generally ready for consumption by users of the application. However, we can look at storing the raw data in addition to processed data for archival purposes. We can control access to ‘raw’ data to administrators alone.
- Processing
This is the phase where we run our descriptive or predictive statistical algorithms. This could also be where the data is opened up for ad-hoc or exploratory analysis.
Presentation: Given the high latency nature of processing, we would like to extract commonly used details for consumption by applications that have low latency requirements. This is usually related to meeting standard reporting requirements, etc. We need to decide what our integration requirements are and how to ensure the data is available for that consumption.
- Archival
This generally is related to compressing and offloading data, for purposes of compliance. This is important to consider as it does drive requirements for capacity planning etc.
Having seen the common usage pattern, this section describes an example architecture, which points out the different set of systems and where they can fit in a BigData solution.
The first step in an analytical process is generally to move from sparse data sources to a co-existing analytical store. Access patterns in an analytical processing system are different from traditional systems. The data is analyzed as a whole set and involves bulk-processing, de-normalized forms etc. and hence analytical stores have special characteristics. Stores can be big data file systems like HDFS, NoSQL databases like HBase, or enterprise traditional data warehouses. This is where data ingestion systems come into picture, which handles the movement of data into analytical stores. The Processing layer is split into two, processing and query. The processing layer performs data manipulation using some sort of ETL process or use of various statistical/machine-learning algorithms, which consume data to derive some insight. The query layer involves data arrangement and structuring so that it can be easily retrieved later. In general, this layer provides a SQL interface on top of the underlying data/analytical stores. The output of the processing layer could be put back to the stores for more efficient query in the presentation layer. The following figure shows the schematic of the various components in analytical big data solution and how they assemble together.
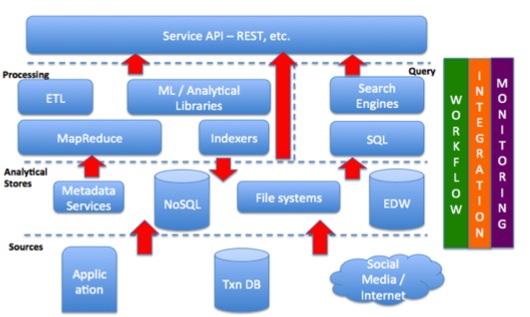
With the understanding of generic and commonly seen architecture, we present Fladoozive and its components.
How did we get the data in?
One of the first problems we had to solve was to get the data into a place where we can use it for further processing. Having reasoned out to choose Hadoop’s HDFS as the analytical store, the idea was to use a component which can work in a reliable fashion but could also write to HDFS. We evaluated Scribe, a server for aggregating log data streamed in real time from a large number of servers, designed to be scalable, extensible without client-side modification, and robust to failure of the network or any specific machine. Facebook developed Scribe and they have used the same for all their log related problems. The problem we faced with Scribe was with the time consuming nature of debugging the scribe instance to write to HDFS. Without the official/community support and the less active development implies that maintaining a system built using this would have costly implication in the future if there is a need to upscale or there is a upgrade in other components. Other options available for large-scale distributed data ingestion component were Twitter Storm and Apache Flume (Couple of other proprietary and OSS implementation we haven’t discussed here). Storm is a distributed realtime computation system. Similar to how Hadoop provides a set of general primitives for doing batch processing, Storm provides a set of general primitives for doing realtime computation. The kind of data requirement we had in hand was not so inclined towards primitive realtime computation/data processing and would have seemed an overkill to use storm. The primary task of the ingest component was to provide a scalable mechanism to get data into HDFS. Apache Flume (Flume-NG is the new generation version of the original apache flume project and the we mean Flume-NG when we refer to flume) is a distributed, reliable, and available service for efficiently collecting, aggregating, and moving large amounts of log data. It has a simple and flexible architecture based on streaming data flows and provides many reliable, failover, recovery mechanism, which makes it robust and fault tolerant.
Flume servers were configured to receive Avro (Data serialization mechanism) messages from different clients emitting logs. The Flume server provides certain handles, which can be used to tune the system for desired operational requirements. It has a concept of channel, which can be used as fast intermediate queues before writing to the sink. MemoryChannel provides high throughput but loses data in the event of a crash or loss of power. Relatively more persistent channel is a FileChannel. The goal of FileChannel is to provide a reliable high throughput channel. FileChannel guarantees that when a transaction is committed, no data will be lost due to a subsequent crash or loss of power. Channels have different configuration capabilities to control the velocity of flow and the latency needs before writing to HDFS or another set of flume servers (in general, sink). In a complex and massively high throughput environment the flume servers can be arranged in a layered architecture to add more reliability and throttling mechanism and hence control the data flow. Flume can write to a set of sinks HDFS, AVRO, File, HBASE, ElasticSearch etc. In our case the final Flume sink wrote to HDFS and was configured for various write characteristics like rollInterval, size, count etc.
What did we do with the data?
Data Organization and Processing
Data life cycle involves storing data at three different stages in processing/analysis. The raw ingested data forms the initial stage in the lifecycle where it is primarily stored for further analysis. This stage may involve creation of suitable directory structure, which would help in making temporal decision either for computational purposes, compliance or monitoring purposes. The next stage is the processing/processed stage where the data after some processing is put in a directory structure, which is more suitable for the usage pattern. Incase the data is used to get timely reports/insights, and uses a Hadoop based querying solution then it’s a good idea to partition the data which will aid in modeling this better. The third stage is the archival stage where the data is stored in a compressed format for any further requirements, be it legal or verification etc.
In the system we developed, the lowest level of directory had all the logs for a given day. The logs themselves did not have enough (rather any) meta data to suggest they should goto a certain directory. This is where Flume’s source interceptors came to rescue. TimeStamp interceptor was used to first tag the incoming log and use this tag information to create a directory on HDFS. This facility/feature was provided by Flume’s HDFS sink dynamic path, which had the ability to parse interceptor/header information. Flume ingested data formed the raw stage of the lifecycle. Processing stage involved running mapreduce Jobs to perform the data cleansing and preparation or in the BI world it’s equivalent to performing ETL (Extract Transform and Load) operations. The resultant data was written on to a processed section with a similar directory structure as the raw data. The important factor here was that the chosen directory structure aided in creating Hive partitions for querying which we will talk about in the next sub-section.
Now that there is data on HDFS, How do you make it query-able?
Hive is a data warehouse system for Hadoop that facilitates easy data summarization, ad-hoc queries, and the analysis of large datasets stored on HDFS or the kinds. It provides a SQL like querying mechanism called Hive Query Language (HQL), which in turn is converted to a set of map reduce jobs and run by Hive to fetch the results. Hive maintains Hive table meta information in an external database (can be configured to point to Mysql databse, and we used external MySql db as the metastore). The challenge was to make Hive point to the newly available data as and when they are available after processing. This was taken care by adding the processed directory, as a new Hive partition after the processing map-reduce job was complete. Hive partitions work in a similar fashion as that of a RDBMS partition. Hive partitions can point to directories on HDFS and the directory structure can act as column partition values. Adding a partition implies a new entry in the Hive metastore and thus the new data can be made available for querying in Hive. Hive provides a sql like interface which can be used to perform exploratory analysis and run queries.
Querying capability, check ✓ Now, How do I make this analyzed data available to my traditional reporting application accessing information from RDBMS?
Exporting data from HDFS to External RDBMS is a common problem faced by Analytical stores and solutions. We have used Sqoop, a tool designed for efficiently transferring bulk data between Apache Hadoop and structured datastores such as relational databases. Sqoop can work with Mysql out of the box using the JDBC driver to connect to it. It also has support to take in a custom driver and connect to other proprietary RDBMS e.g. Microsoft’s SQLServer. In Fladoozive, the output of a Hive query was written on to HDFS and Sqoop was used to pick up this result and populate a remote RDBMS table. In this way, a system, which reports insights on a timely manner can still work on the traditional RDBMS without having to move to a distributed paradigm as a set of operation can perform computation in batches on a distributed eco-system like Hadoop and dump the final output to RDBMS.
Talking about automation and integration of so many systems, How can I manage these workflows using just one tool/framework?
The typical process, which we emphasized in the previous sections, involves performing tasks in an orderly fashion, as a workflow. There are a bunch of options available to manage Hadoop related workflows e.g. Cascading, Oozie, Azkaban, Hamake etc. In our case the requirement was to be able to include actions from different components of Hadoop Ecosystem as part of one workflow. Cascading provides Java apis to schedule several kinds of jobs, Azkaban and Hamake have other ways to express a workflow but on the other hand support very few kinds of Hadoop related jobs. Given our requirement to integrate Mapreduce, Hive, Sqoop actions in a workflow, Oozie seemed a better fit as it supported most of these jobs out of the box. Oozie is a workflow scheduler system built to manage Hadoop Jobs. It is integrated with the rest of the Hadoop stack supporting several types of Hadoop jobs out of the box (such as Java map-reduce, Streaming map-reduce, Pig, Hive, Sqoop and Distcp) as well as system specific jobs (such as Java programs and shell scripts). Once Oozie is configured to work with Hive, Sqoop it can perform a task pertaining to these tools. Configuring Oozie to work with them at times simply involves making the java package jars available in the execution path on HDFS. Oozie workflow can simplify the complexity involved in liking multiple Hadoop Map Reduce jobs and also can aid in the easy execution of other tools in the Hadoop Ecosystem which come as part of a particular data work flow.
We had two sets of Oozie workflow:
- Pure ETL and processing, which would transform data into query-able format E.g. add Hive partition after some processing.
- Jobs of query and export nature.
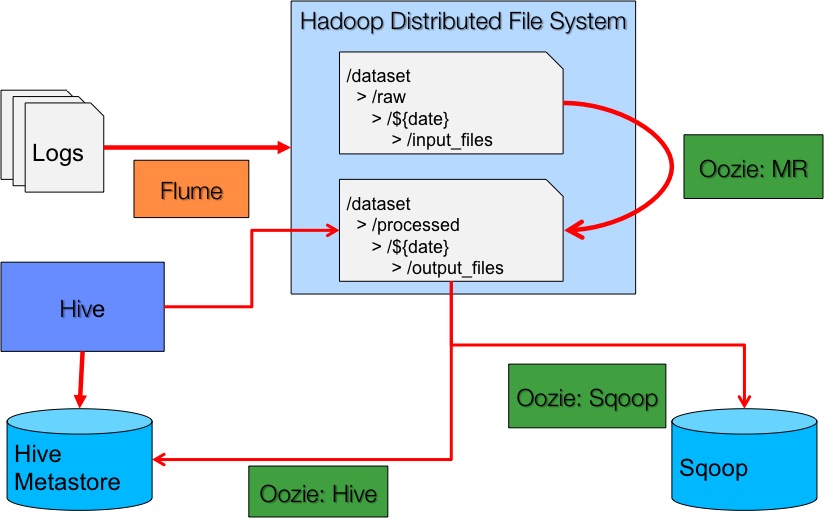
Building the system gave us opportunity to understand better the distributed computing paradigm at an enterprise scale and what it takes to deal with the obstacles encountered along the way.
Conclusions
It was satisfying to see a system getting built over the course of a week and a half and just work. All of our choices were Open Source applications and for organizations looking for a custom solution due to privacy requirements or extensibility of use, at lower cost and wide community support, we would definitely recommend use of Hadoop Eco-System. There are certain areas in which the whole eco-system can improve. There are already signs that the community is doing everything there is to get to that stage.
There is a solution available for any specific problem, which one can think of. The ecosystem is evolving big and strong, there are already systems adopting newer features in Hadoop (YARN) and leveraging this to build more sophisticated systems which could work on the Hadoop stack. One of such discrete example is the evolution of SQL like solution that use HDFS. It is not long that Hadoop and its Eco-System becomes the De-Facto Standard for any analytical/Big Data solution.
Highlighting Important Lessons Learnt
Always check for version compatibility before proceeding: When integrating different components, always check if the versions of the components can co-exist. Tiny peril of using OSS components but on the contrary, public support forums come to the rescue in no time.
Be prepared to get your hands dirty with the source code if necessary (which is rare) the component is in its very nascent stage and there is relatively less documentation around certain not fully evolved feature.
[Originally Posted on Blogspot]